Clinical Research Data Integration
Architecture, Technologies, Case Studies
In healthcare IT since 2005, ScienceSoft helps academic institutions, CROs, pharmaceutical and biotech companies build clinical data integration software that ingests data in many formats, automates data processing and QC, and offers powerful tools for clinical research analytics and data mining.
Over Half of Current Clinical Trials Use Six or More External Data Sources
A recent eClinical’s survey of small, mid-sized, and large clinical trial sponsors showed that 65% of respondents used six or more external data sources within their studies, and 29% used more than 10.
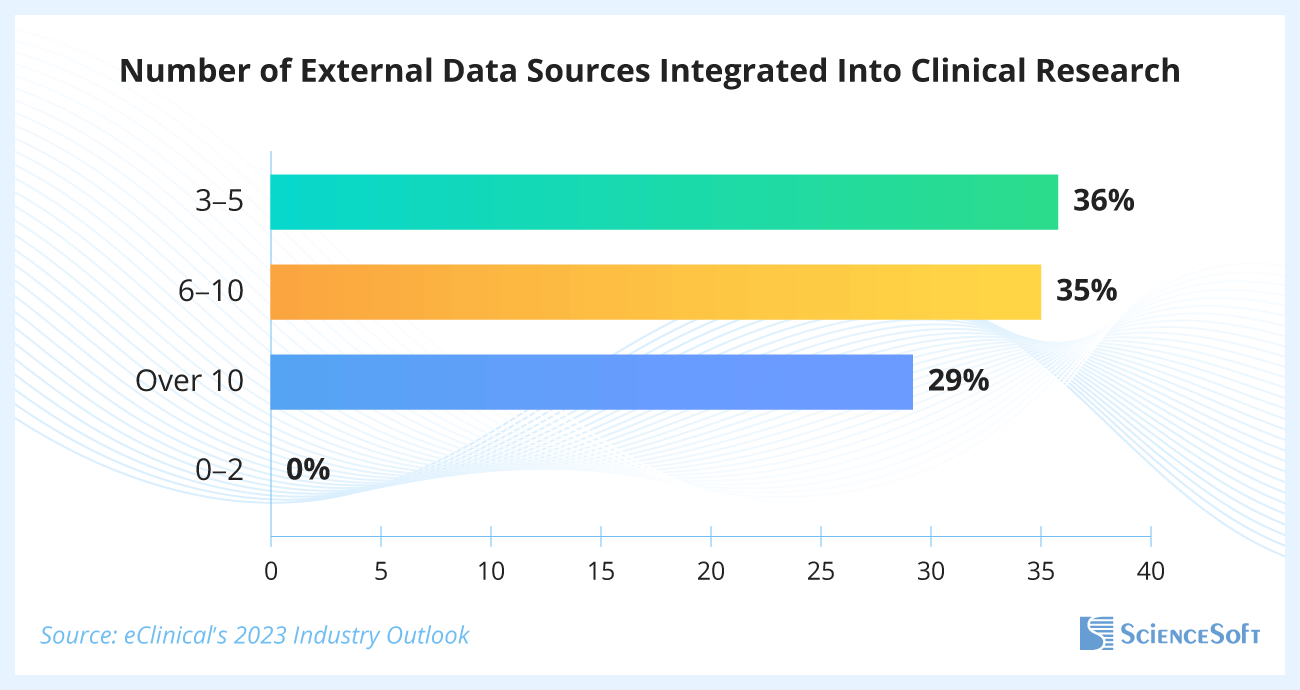
Surveyed specialists shared that challenges with accommodating the growing number of data types, data standardization, and reconciliation result in the following pressing issues:
- 30% of respondents reported prolonged study timelines, specifically the inability to meet target timeframes from the last patient's last visit to database lock and from database lock to final TLFs (tables, listings, and figures packages’ finalization).
- 30% encountered problems with data quality.
- 19% faced insufficient flexibility of existing software for the proper integration of the necessary data.
Notably, the experts surveyed pointed to effective data management automation as the biopharmaceutical industry's number one priority (36%), placing it above other trends like decentralized clinical trials (11%) and risk-based practices (12%), due to the pressure to speed up cycle times.
Clinical Data Integration Technology at a Glance
Clinical data integration technology merges, standardizes, and validates heterogeneous clinical data from multiple sources, making it fully visible, shareable, and suitable for automated management and analytics, including via big data and ML/AI tools.
In clinical practice, data integration enables:
Patient data exchange
Record exchange with partner diagnostic centers, hospitals, outpatient clinics, and insurers, ensuring care continuity.
Solving inefficiencies in physicians' performance
E.g., prolonged hospitalizations, unnecessary tests, and claim rejections, caused by missing patient data.
Monitoring and improving care quality
Specifically, to achieve the quality targets established by the Centers for Medicare and Medicaid Services.
Healthcare organizations often use clinical data integration tools as part of:
- Health information exchange (HIE) software that enables interaction with partners and insurers for service delivery and cost coverage.
- A healthcare data warehouse that, in addition to clinical data, also integrates the company's operational and financial details for tracking, analysis, and guiding both clinical and business decisions.
In biomedical research, clinical data integration makes it possible to:
Leverage diverse patient data sources
E.g., output from wearables, genomic data from biobanks, or longitudinal real-world patient data from health systems.
Automate data processing and quality control
of extensive and diverse research data, shortening trial timelines and lowering expenses.
Use cutting-edge analytics on large data sets
E.g., AI-driven pattern recognition, predictive modeling, or deep learning for genomic data interpretation.
Contract research organizations (CROs) and R&D pharmaceutical and biotech companies typically use clinical data integration tech in clinical data management systems.
Sample Architecture of a Clinical Data Integration Solution for a CRO
Clinical data is typically integrated using a process known as Extract, Transform, and Load (ETL). Below, ScienceSoft's principal software architects explain the data flows, data transformation stages, and key architectural modules involved in this process. For illustration purposes, we are using a sample solution intended for clinical research organizations.
For clinical data integration, we usually opt for the medallion architecture, which provides an incremental improvement in data quality as it moves from the bronze data layer through silver to gold. The bronze layer ingests and structures raw data. The silver layer filters, cleans, and enriches it. The gold layer delivers clean, continuously updated data to users for solving their tasks.
A major benefit of the layered structure is that it gives full traceability of research data. You can track every step of its transformation and view every document version, which is essential for both researchers and auditors to ensure reproducibility and regulatory compliance.
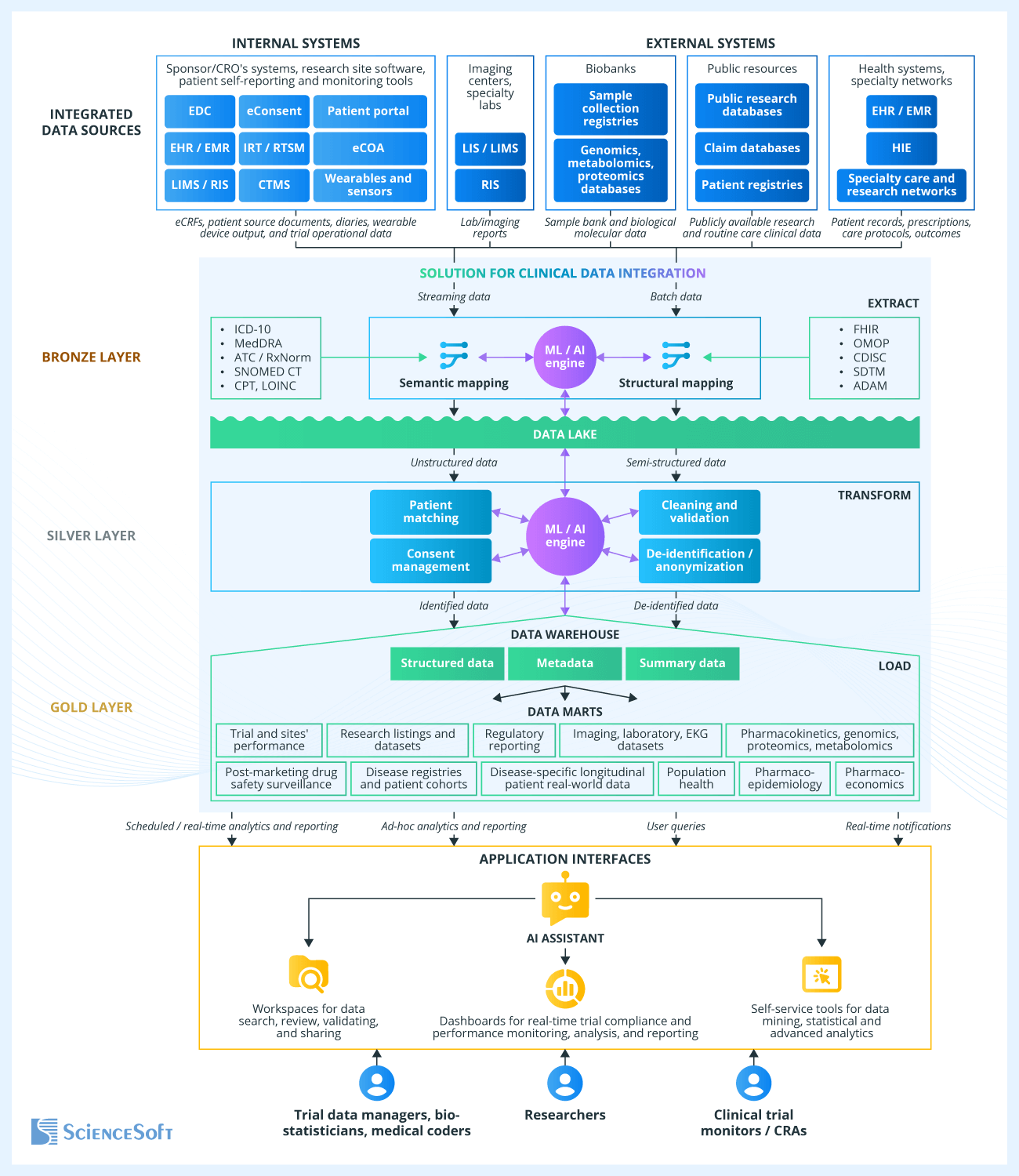
Data extraction
Clinical research typically requires data ingestion from both internal systems (such as software for capturing patient data, CTMS, and IRT) and external sources (such as specialty labs, biobanks, and real-world patient data accumulated in EHR by healthcare providers).
The integration solution may pull clinical data from these sources through FHIR REST APIs (for EHR integration, USCDI-aligned content exposed via certified APIs under the 21st Century Cures Act). Other options include direct database access (e.g., for internal software), file export, HL7 v2/v3 messages (for legacy systems), or CCDA documents (for patient visit summaries and discharge notes). Beyond structured data and texts, source data types include waveforms, medical images, audio and video files, and outputs from medical sensors and wearable devices.
Source data can arrive as a real-time stream (e.g., sensor readings from wearable or implantable medical devices), on demand (e.g., eCRFs from EDC or patient diaries from eCOA get ingested as they are completed), or in scheduled batches (e.g., lab results or public research datasets). Costly real-time data streaming is typically reserved for time-critical use cases, such as remote patient monitoring, where delays in processing could pose risks to patient safety.
Semantic and structural mapping
The first step in the data transformation process involves semantic mapping between the source terminologies and multiple national and international terminology standards, such as ICD-10 for diagnoses, ATC or RxNorm for medications, CPT for procedures, SNOMED CT for conditions, LOINC for lab results, etc. For example, you could use semantic mapping to standardize the adverse event terms according to the Medical Dictionary for Regulatory Activities (MedDRA).
Unified terminology facilitates the next stage in clinical data transformation — structural mapping. This process identifies field names, formats, value types, and relations in the source data (e.g., for visits, procedures, and medications) and converts them to a unified format according to a standardized data model. Common examples of such data models include those designed by Observational Medical Outcomes Partnership (OMOP) and Clinical Data Interchange Standards Consortium (CDISC), the Study Data Tabulation Model (SDTM), and more.
The structural mapping module also handles unifying units of measurement and dates and tagging data elements with semantic metadata for easier search and interpretation.
Ingested data goes into a data lake that serves as a storage for raw (unstructured and semi-structured) clinical data.
Data cleaning and validation
This clinical data transformation module performs automatic inspection for invalid and missing data, out-of-range values, discrepancies, and outliers. It also evaluates the quality of earlier processing steps by reviewing the completeness of standardized data versus the sources, validating conformance to the target data model, and checking the percentage of data successfully mapped to target vocabularies.
Following the set rules, the solution logs inspection results, flags unmapped fields or values, auto-corrects or flags quality flaws, and alerts data managers if data quality goes below the benchmark level.
Patient matching and consent management
The patient matching module identifies and links together records ingested from different sources that belong to the same patients. For example, it can merge genomic data collected by the cancer care network with the electronic health records (EHR) of the corresponding patients stored in other healthcare organizations to examine potential tumor risk factors.
In addition, the software monitors and tags the personal and health data processing types and disclosure purposes to which patients have given consent, so that compliance can be enforced during clinical data access or use. The software also keeps track of any updates or withdrawals of consent made by patients.
De-identification and anonymization
The de-identification module removes from clinical data all fields that can directly identify an individual, such as name, phone number, and email. Usually, this involves assigning patients new random identifiers, pseudo-IDs — in that case, it’s called pseudonymization. This allows maintaining an encrypted link between real and pseudo IDs, which is often required for updating records and performing analysis.
For example, this allows patient records to be automatically updated with new treatment results coming from their healthcare providers. If needed, researchers can trace any record back to the original document to rule out errors or study what influences the drug’s effectiveness.
In the case of full anonymization, no link is preserved between the patient’s identity and their data, and extra steps are taken to prevent identity recovery through data mining, such as combining region, age, diagnosis, and medical history details. The software can apply several methods of clinical data de-identification, so that each dataset includes only the minimum amount of PHI needed to achieve the research goals.
Data loading, consolidating, and storage
Transformed, cleaned, and highly structured clinical data is uploaded into the clinical data warehouse, where it is stored along with a structured metadata repository and data summaries.
Metadata contains detailed data descriptions, their origin, and change logs. For example, it allows for semantic search of patients by the type of procedure performed, tracking these records back to the source, receiving automatic updates on outcomes, checking procedure protocol versions, and data quality auditing.
Data summaries provide a high-level overview of the clinical data, including aggregated statistics. For instance, it helps researchers promptly retrieve summarized data on public health in different regions, along with automatically calculated statistical and epidemiological indicators.
Structured clinical data is also sorted and integrated into data marts — multidimensional datasets required for specific research needs. For example, a data warehouse may store datasets containing pharmacokinetic data for different patient subgroups, drug safety data collected during the study and extracted from EHR, longitudinal real-world data of patients with specific conditions, etc.
ML/AI-driven data processing
Advanced software for clinical data integration usually includes an ML/AI engine that uses natural language processing (NLP), deep learning, and other techniques to improve data transformation efficiency across all functioning modules of the solution:
- Semantic mapping: Recognizing semantic structures in text notes, audio and video recordings, attachments, and images; identifying the meaning of source terms based on context and finding the best match for them in the standard vocabulary.
- Structural mapping: Identifying source record formats and the data field locations, matching data to the target data model, structuring text notes and media files, and performing semantic data annotation.
- Data cleaning: Reviewing data in context and using trend modeling to uncover anomalies, outliers, and data that seems unrealistic. Identifying potential error sources, suggesting corrections, and offering substitution options for missing data.
- Patient matching: Confirming patient identity for ingested data, checking for matched patient EHR in all available systems and databases.
- De-identification: Detecting and deleting identifiers from text notes, attachments, media content, and medical images.
- Data mart extraction: Filtering and sorting structured data based on context, matching records ingested from different sources to build multidimensional datasets.
Data serving
In the clinical data warehouse, highly structured data is stored in a way that maximally simplifies its search, viewing, sharing, analysis, and reporting. Users access the data through their interactive dashboards, which visualize automated reports and include tools for data exploration, AI-assisted querying and analytics.
For example, data managers can review the trial data with marked quality defects, trace the source of errors, and identify their common causes. In the same way, researchers conducting non-interventional studies can use a search tool to extract patient records that meet specific criteria, along with advanced tools for data mining and AI analytics.
Why Entrust Your Clinical Data Integration Initiative to ScienceSoft
- Since 2005 in healthcare software development.
- 150+ successful projects in the domain.
- Since 1989 in data analytics, data science, and AI.
- Expertise in healthcare data exchange standards (FHIR, HL7 v2/v3, CCDA, USCDI), terminology and coding standards (ICD-10, CPT, LOINC, SNOMED CT, RxNorm), and medical imaging standards (DICOM).
- Experience in meeting GCP, FDA, HIPAA, GDPR, and 21st Century Cures Act requirements.
- Project managers, principal architects, consultants, and software engineers with 5–20 years of experience and a background in clinical research software projects.
Our awards, recognitions, and certifications
Featured among Healthcare IT Services Leaders in the 2022 and 2024 SPARK Matrix
Recognized for Healthcare Technology Leadership by Frost & Sullivan in 2023 and 2025
Named among America’s Fastest-Growing Companies by Financial Times, 4 years in a row

Top Healthcare IT Developer and Advisor by Black Book™ survey 2023
Four-time finalist across HTN Awards programs

Named to The Healthcare Technology Report’s Top 25 Healthcare Software Companies of 2025

HIMSS Gold member advancing digital healthcare
ISO 13485-certified quality management system
ISO 27001-certified security management system












